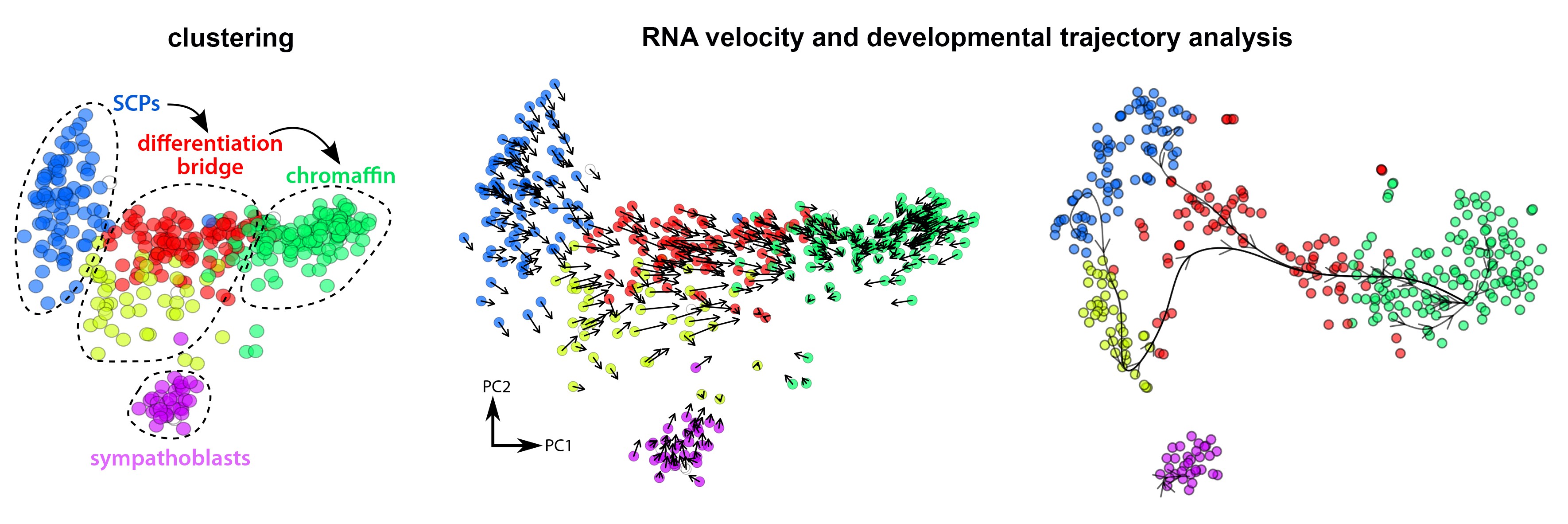
Our expertise lies in applying high-resolution single-cell RNA sequencing (scRNA-seq) and integrative multiomic approaches to study cell fate decisions, lineage trajectories, and tissue regeneration. By combining transcriptomic, epigenomic, and spatial omics data, we decode the molecular mechanisms underlying neural crest development, neuroglial interactions, and disease progression. Our lab develops advanced computational pipelines for data analysis, ensuring deep insights into cellular heterogeneity across various biological systems.
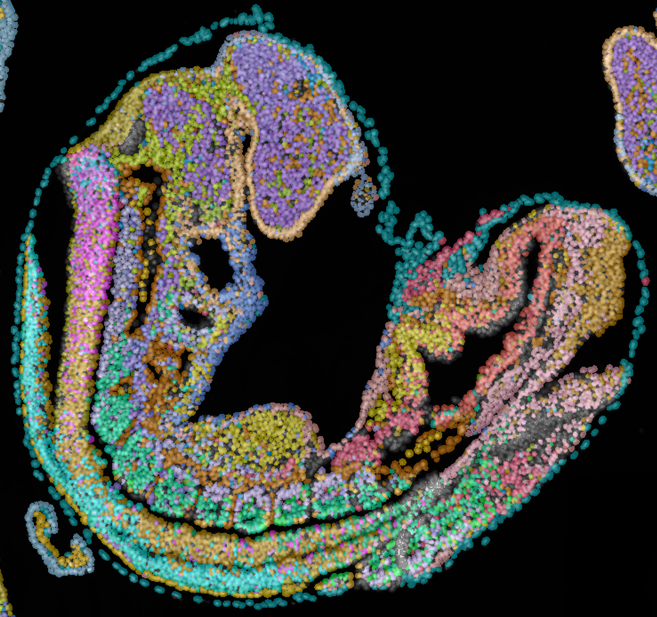
Adameyko Lab specializes in spatial transcriptomics, leveraging cutting-edge technologies to map gene expression with unprecedented resolution. Our expertise includes MERFISH, Visium HD, Xenium 5K, and Stereo-seq, enabling precise spatial profiling of cellular ecosystems across diverse tissues and developmental stages. By integrating these powerful approaches, we uncover cellular organization, microenvironmental interactions, and lineage dynamics in complex biological systems. Our research combines high-throughput imaging, spatially resolved RNA sequencing, and computational modeling to decode fundamental processes in neural crest biology, neurodevelopment, and regenerative medicine.
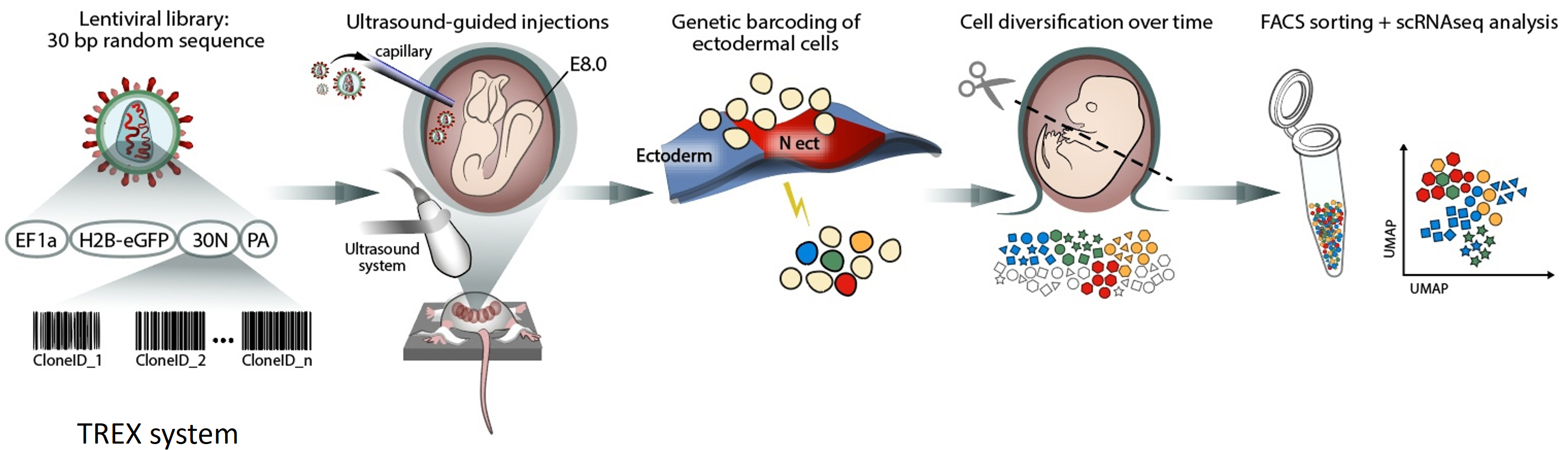
Adameyko Lab innovates in the field of high-resolution clonal analysis, employing cutting-edge clonal barcoding and DNA scarring approaches to trace cell lineage dynamics with unparalleled precision. By integrating these technologies with single-cell transcriptomics and spatial omics, we reconstruct developmental trajectories, stem cell behavior, and tissue regeneration at a clonal level. Through innovative experimental and computational pipelines, we provide deep insights into how individual cells contribute to tissue architecture and function over time.
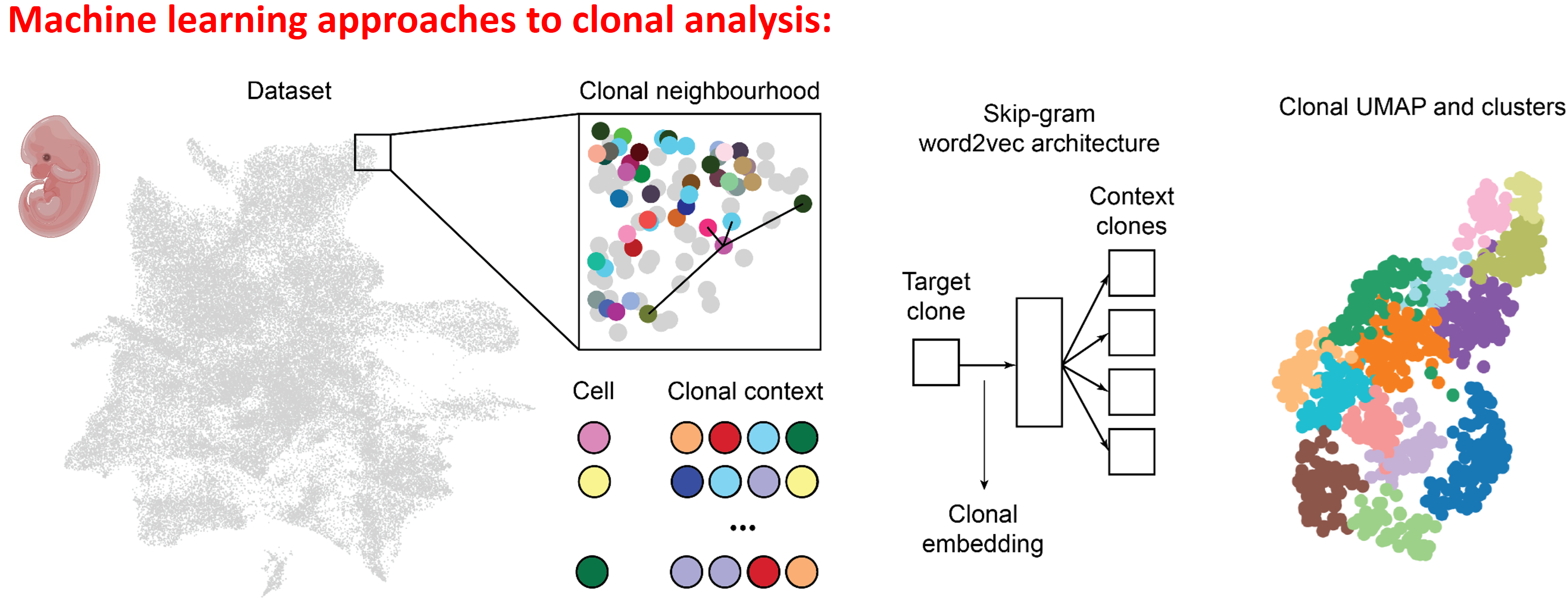
Adameyko Lab integrates machine learning with clonal analysis and genomic regulatory landscape mapping to uncover the principles of cell fate decisions and tissue development. By leveraging ATAC-seq, scCUT&Tag, and scCUT&RUN, we decode chromatin accessibility, transcription factor dynamics, and epigenetic regulation at single-cell resolution. Our advanced computational frameworks apply deep learning and probabilistic modeling to reconstruct clonal lineage relationships, predict regulatory networks, and identify key epigenetic drivers of cellular transitions. By combining these approaches with single-cell transcriptomics and spatial omics, we generate high-resolution, predictive models of development, regeneration, and disease progression.
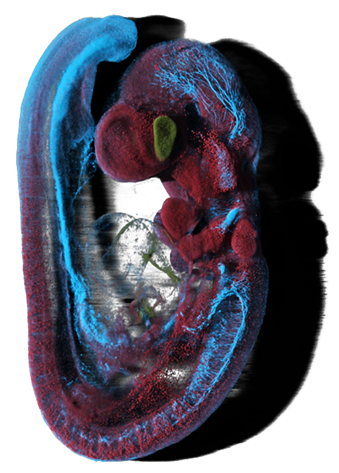
Adameyko Lab uses advanced 3D microscopy, enabling high-resolution visualization and quantitative assessment of complex biological structures. We specialize in spinning disk microscopy and light sheet microscopy, capturing dynamic cellular processes in developing and regenerating tissues with exceptional depth and clarity. Our expertise includes 3D reconstructions, image segmentation, and data analysis using IMARIS. By integrating these approaches with single-cell transcriptomics and lineage tracing, we uncover morphogenetic patterns, cell-cell interactions, and tissue organization in great detail.
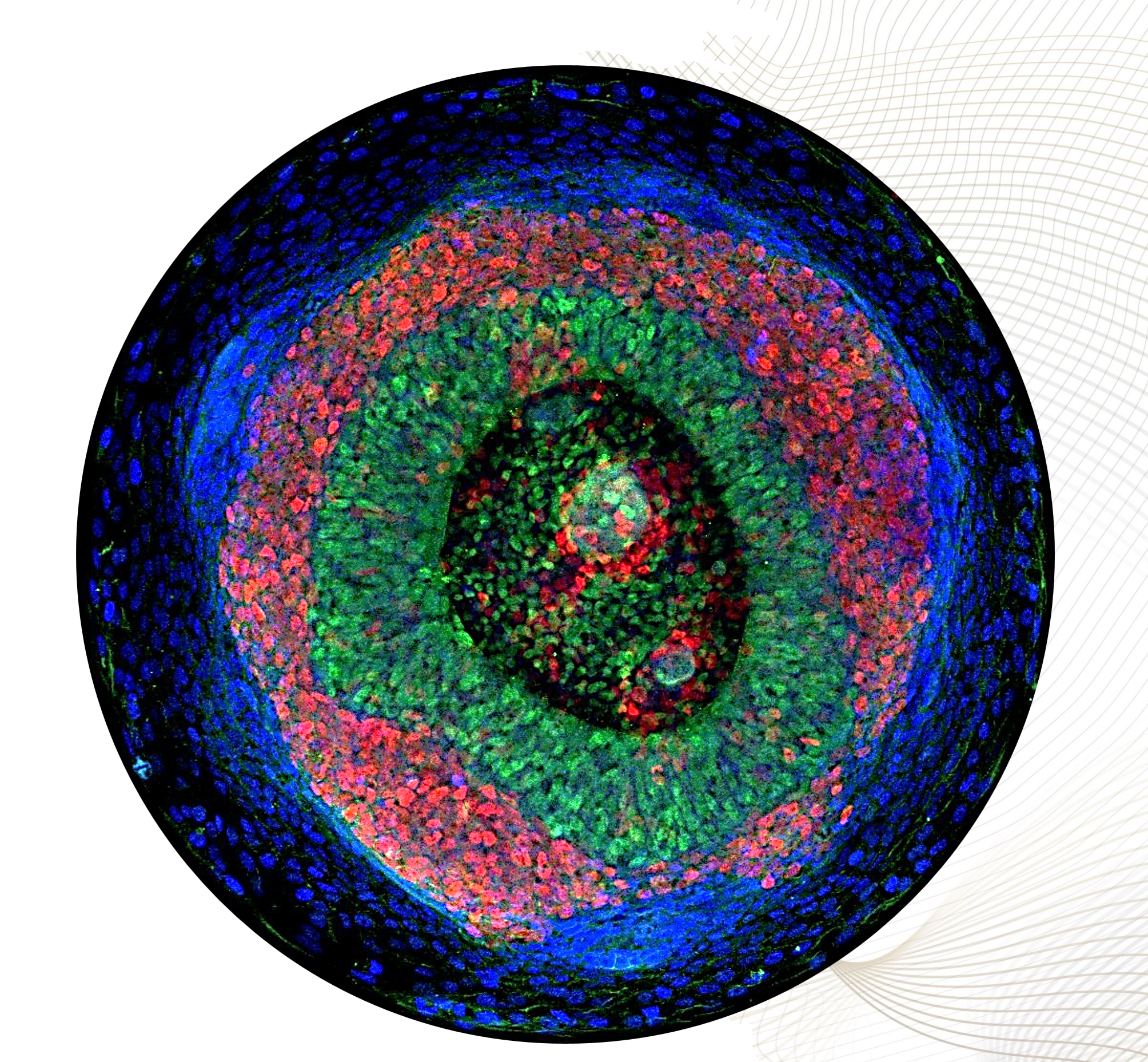
Adameyko Lab invests efforts in growing and studying organoids and assembloids to model the development, interactions, and diseases of the central and peripheral nervous systems. By leveraging cutting-edge stem cell differentiation protocols, bioengineering approaches, and single-cell multiomics, we generate functional neural tissues that mimic in vivo complexity. Our expertise includes co-culturing CNS and PNS organoids, enabling us to investigate neuroglial interactions, neural crest contributions, and axon guidance in a physiologically relevant context. By integrating spatial transcriptomics, live imaging, and machine learning-based analysis, we uncover fundamental mechanisms of neurodevelopment, regeneration, and neurological disorders
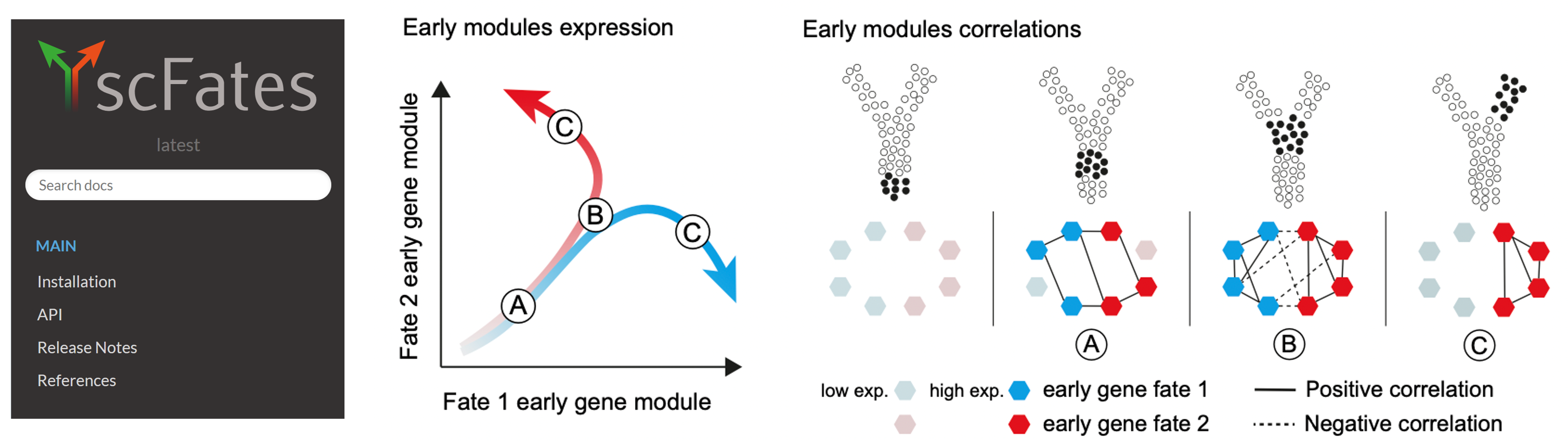
Adameyko Lab is profocient in cutting-edge computational methods for cell fate prediction and gene regulatory network (GRN) reconstruction from single-cell data. Using tools like scFates, we apply advanced trajectory inference and probabilistic modeling to uncover cell state transitions and lineage dynamics in complex biological systems. Our expertise extends to GRN reconstruction with innovative platforms such as CellOracle and Regvelo, allowing us to predict transcriptional regulators, epigenetic influences, and gene interactions driving cell fate decisions.





