Our work combines both invertebrate and vertebrate model organisms, exploiting the versatility of Drosophila genetics, the detail knowledge available on the structure and function of the rodent nervous system, and our unique access to animals that adapted to extreme environments and show evolutionarily unique features, like longevity and cancer resistance in naked mole rats.
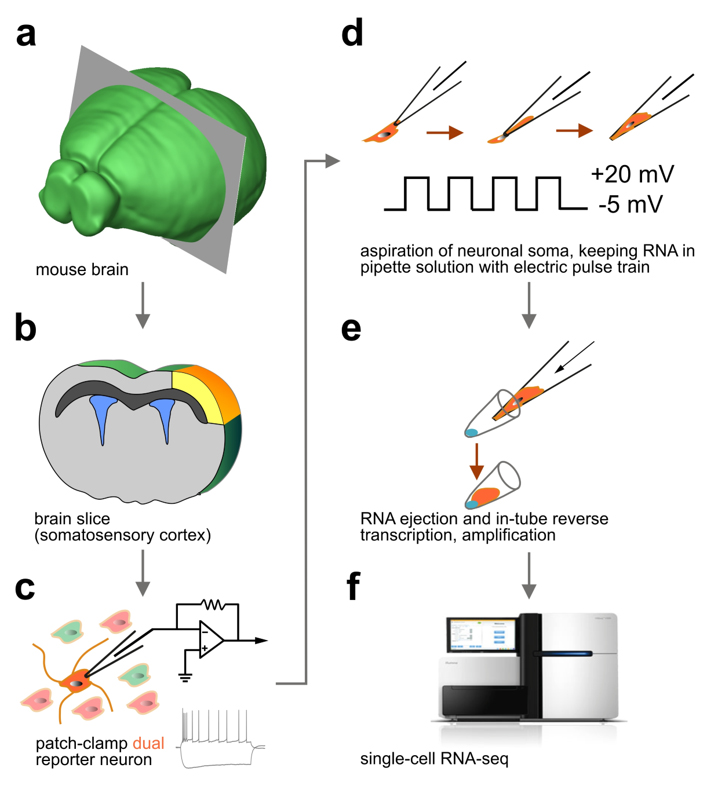
We were first to develop ‘Patch-seq’, the most advanced cellular profiling workflow widely used in neuroscience to classify neurons (Fuzik et al., Nature Biotechnology, 2016). Patch-seq takes advantage of the genetic tagging of cell types, their recording by slice electrophysiology (or even in vivo), intracellular biocytin loading for post-hoc neuroanatomy, and single-cell RNA-seq for molecular profiling.

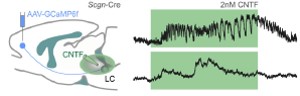
We routinely use ex vivo (slice) electrophysiology to address fundamental features of neuronal biophysics, as well as of synaptic communication in neuronal networks. Recently, we have expanded our repertoire to calcium imaging using genetic sensors (GCamPs), and GRABs. We have muticlamp systems allowing the routine analysis of 3-4 cells at any given time.

We have long-standing interest in protein biochemistry, particularly in drug-induced changes during brain development, and its modeling in cell lines, primary neurons, and co-culture systems. For protein determination, we have acquired an Amersham automated WB system as beta test site (GE healthcare) and used total protein normalization as a precise tool for comparisons. Moreover, we have relied on mass spectrometry, largely in collaboration with the Lubec group (PMU Salzburg) and the Scottish proteomics Core (University of St. Andrews) for our discovery research.

We were first in Austria to acquire a Lightsheet Z1 imaging system from Zeiss, which has been upgraded as our work-horse ever since. Together with our LSM880 Airyscan, LSM710, and ApoTome.2 microscope, Imaris 10.0.1 workstations (for image processing), an imaging suite is available to reach from single molecules to intact neurocircuits in optically-cleared brains.
We are excitedly awaiting a new addition: a ultrafast single-molecule tracking microscope in 2024.

We use Noldus PhenoTyper cages, equipped to monitor sleep/wake cycles, food and fluid intake, spontaneous activity, running-wheel activity and other parameters to screen if genetic manipulations and/or pharmacological interventions affect metabolic and neuroendocrine processes. By having these cages equipped for optogenetic recordings, we can assign particular neuronal populations as determinants of specific behaviors.
Moreover, we use elevated plus maze, open field, object recognition and other tests to screen anxiety and depression-like phenotypes.